Health-GPS

Global Health Policy Simulation model
Global Health Policy Simulation model
| Home | Quick Start | User Guide | Software Architecture | Data Model | Developer Guide | API |
Quick Start
The Health-GPS application provides a Command Line Interface (CLI) and runs on Windows 10 (and newer) and Linux devices. All supported options are provided to the model via a configuration file (JSON format), including population size, intervention scenarios and number of runs. Users are encouraged to start exploring the model by using the included example dataset, changing the provided configuration file, and running the model.
Although the model source code is portable, binaries must be generated for each platforms using respective tools and libraries. The Health-GPS repository provides x64 binaries for Windows and Linux Operating Systems (OS) with very specific runtime requirements.
Windows OS
You may need to install the latest Visual C++ Redistributable on the machine, the application requires the 2022 x64 version to be installed.
- Download the latest release source code and binaries for Windows from the repository.
- Unzip both files content into a local directory of your choice (xxx).
- Open a command terminal, e.g. Windows Terminal, and navigate to the directory used in step 2 (xxx).
- Rename the data source subfolder (healthgps) by removing the version from folder’s name.
- Run:
X:\xxx> .\HealthGPS.Console.exe -f healthgps/example/France.Config.json -s healthgps/datawhere-fgives the configuration file full-name and-sthe path to the root folder of the backend storage respectively. - The default output folder is
C:/healthgps/results/france, but this can be changed in the configuration file(France.Config.json).
Linux OS
You may need to install the latest GCC Compiler in your system, the
application requires GCC 11.1 or newer to be installed.
- Download the latest release source code and binaries for Linux from the repository.
- Unzip both files content into a local directory of your choice (xxx).
- Open a command terminal and navigate to the directory used in step 2 (xxx).
- Rename the data source subfolder (healthgps) by removing the version from folder’s name.
- Run:
user@machine:~/xxx$ ./HealthGPS.Console.exe -f healthgps/example/France.Config.json -s healthgps/datawhere-fgives the configuration file full-name and-sthe path to the root folder of the backend storage respectively. - The default output folder is
$HOME/healthgps/results/france, but this can be changed in the configuration file(France.Config.json).
NOTE: The development datasets provided in this example are limited to 2010-2050 time frame. It is provided for demonstration purpose to showcase the model’s usage, input and output data formats. The backend data storage can be populated with new datasets, the index.json file defines the storage structure and file names, it also stores metadata to identify the data sources and respective limits for validation.
Known Issue: Windows 10 support for VT (Virtual Terminal) / ANSI escape sequences is turned OFF by default, this is required to display colours on console / shell terminals. You can enable this feature manually by editing windows registry keys, however we recommend the use of Windows Terminal, which is a modern terminal application for command-line tools, has no such limitation, and is now distributed as part of the Windows 11 installation.
Results
The current model output format is JSON (JavaScript Object Notation), an open standard file format designed for data interchange in human-readable text. It is language-independent; however, all programming language and major data science tools supports JSON format because they have libraries and functions to read/write JSON structures. To read the model results in R, for example, you need the jsonlite package:
require(jsonlite)
data <- fromJSON(result_filename.json)
View(data)
The above script reads the results data from file and makes the data variable available in R for analysis as shown below, it is equally easy to write a R structure to a JSON string or file.
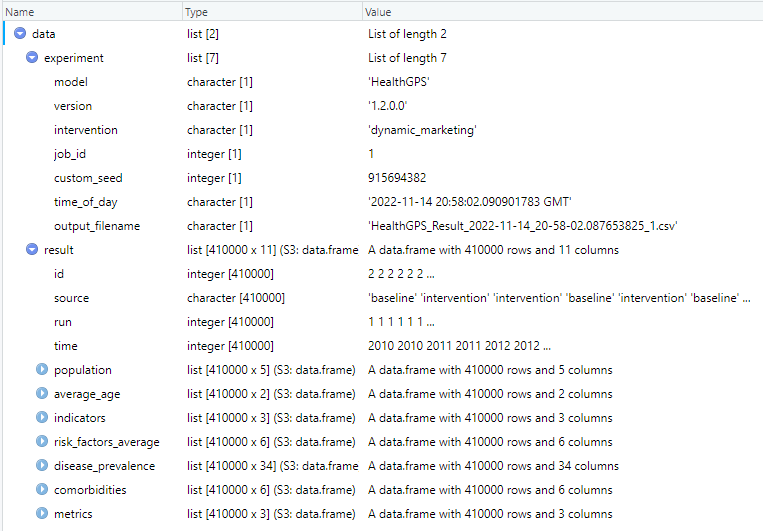 |
|---|
| Health-GPS results in R data frame example |
The results file contains the output of all simulations in the experiment, baseline, and intervention scenarios over one or more runs. The user should not assume data order during analysis of experiments with intervention scenarios, the results are published by both simulations running in parallel asynchronously via messages, the order in which the messages arrive at the destination queue, before being written to file is not guaranteed. A robust method to tabulate the results shown above, is to always group the data by: data.result(source, run, time), to ensure that the analysis algorithms work for both types of simulation experiments. For example, using the results data above in R, the following script will tabulate and plot the experiment’s BMI projection.
require(dplyr)
require(ggplot2)
# create groups frame
groups <- data.frame(data$result$source, data$result$run, data$result$time)
colnames(groups) <- c("scenario", "run", "time")
# create dataset
risk_factor <- "BMI"
sim_data <- cbind(groups, data$result$risk_factors_average[[risk_factor]])
# pivot data
info <- sim_data %>% group_by(scenario, time) %>%
summarise(runs = n(),
avg_male = mean(male, na.rm = TRUE),
sd_male = sd(male, na.rm = TRUE),
avg_female = mean(female, na.rm = TRUE),
sd_female = sd(female, na.rm = TRUE),
.groups = "keep")
# reshape data
df <- data.frame(scenario = info$scenario, time = info$time, runs = info$runs,
bmi = c(info$avg_male, info$avg_female),
sd = c(info$sd_male, info$sd_female),
se = c(info$sd_male / sqrt(info$runs), info$sd_female) / sqrt(info$runs),
gender = c(rep('male', nrow(info)), rep('female', nrow(info))))
# Plot BMI projection
p <- ggplot(data=df, aes(x=time, y=bmi, group=interaction(scenario, gender))) +
geom_line(size=0.6, aes(linetype=scenario, color=gender)) + theme_light() +
scale_linetype_manual(values=c("baseline"="solid","intervention"="longdash")) +
scale_color_manual(values=c("male"="blue","female"="red")) +
scale_x_continuous(breaks = pretty(df$time, n = 10)) +
scale_y_continuous(breaks = pretty(df$bmi, n = 10)) +
ggtitle(paste(risk_factor, " projection under two scenarios")) +
xlab("Year") + ylab("Average")
show(p)
 |
|---|
| Experiment BMI projection example |
In a similar manner, the resulting dataset df, can be re-created and expanded to summarise other variables of interest, create results tables and plots to better understand the experiment.